Exploratory analysis involves data cleaning, feature engineering, and data summarizing and trend analysis.
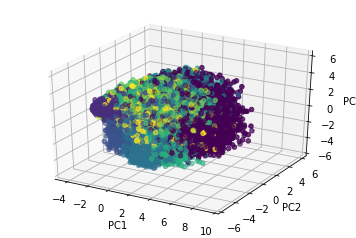
It is common to be faced with a dataset with high dimensionality. This means that there are many possible features (or variables) that can be used in a machine learning algorithm or statistical model. Typically only a small set of these features will actually be informative to a model, and many features will be highly correlated. A large set of features can lead to very long processing time and overfitting. Dumping a large amount of features into a model is a great way to get a garbage in garbage out model. The features need to be reduced and/or transformed before used in developing a model. Exploratory analysis explores the data for meaningful trends, creates meaningful features, reduces dimensionality, and cleans the data so that it is prepared for analysis.
An exploratory analysis can include some of these steps:
- Remove meaningless features from the dataset. If the feature does not have a statistically significant relationship with the response variable y, it is probable y not useful to include it in the model.
- Calculate new features from the dataset.
- Normalize or Standardize data
- Impute missing data
- Apply transformations such as a log transformation, or first order difference.
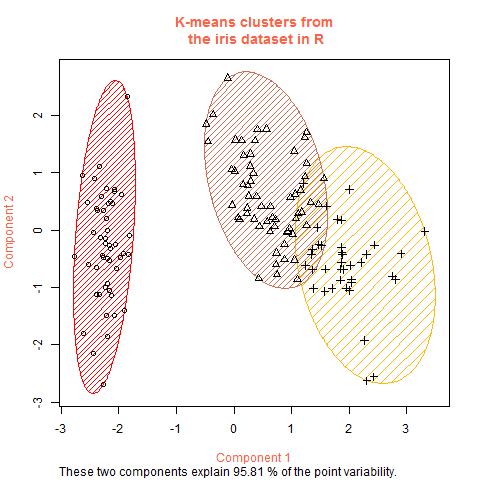
- Apply dimensionality reduction methods such as PCA to decorrelate/summarizes the features
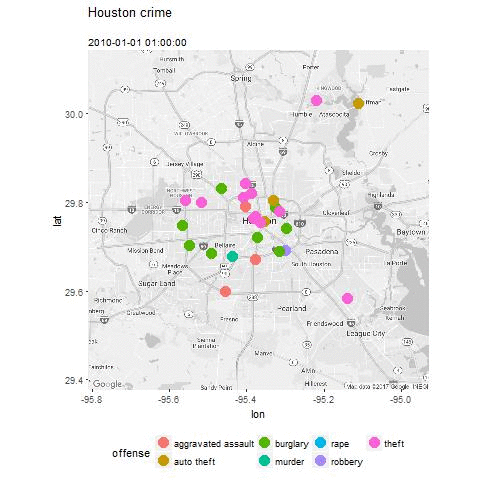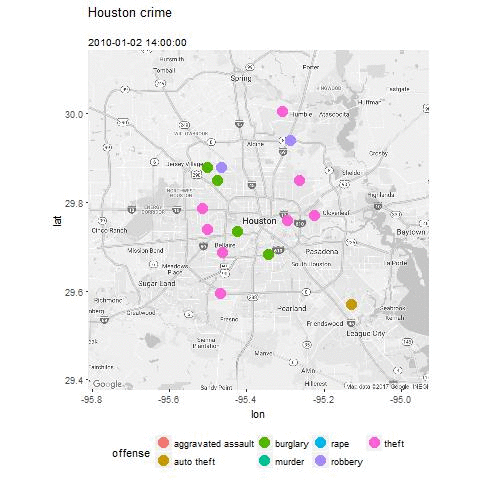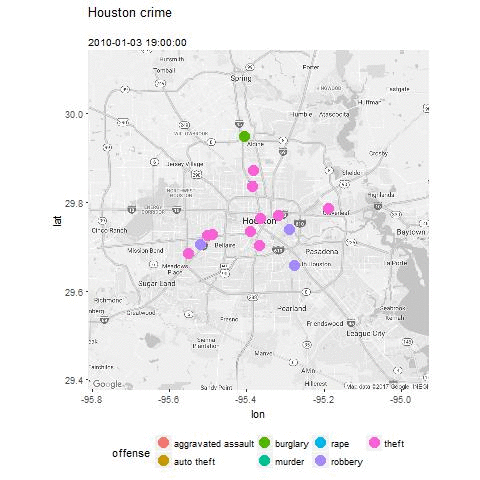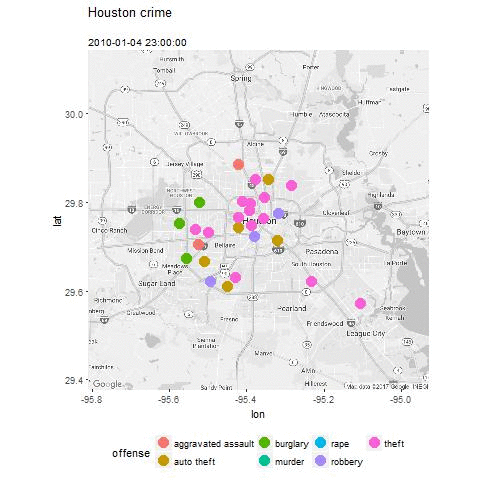
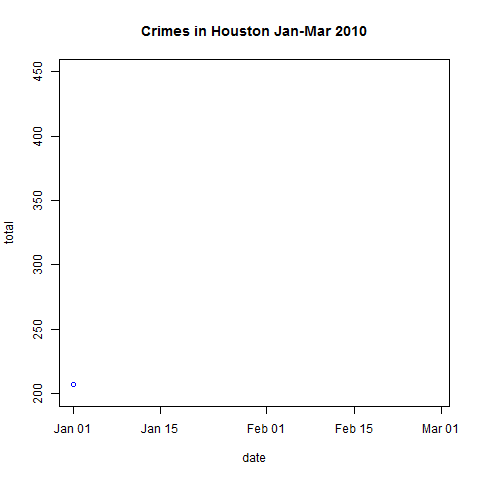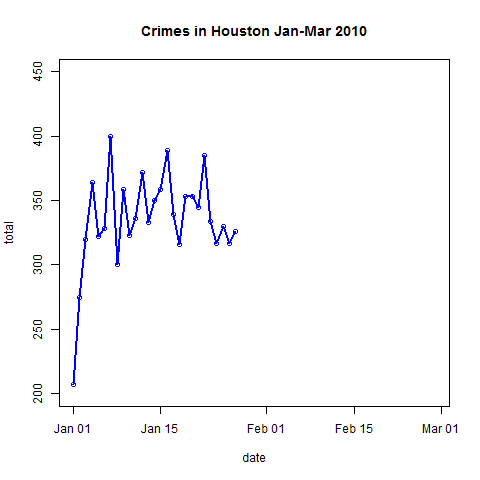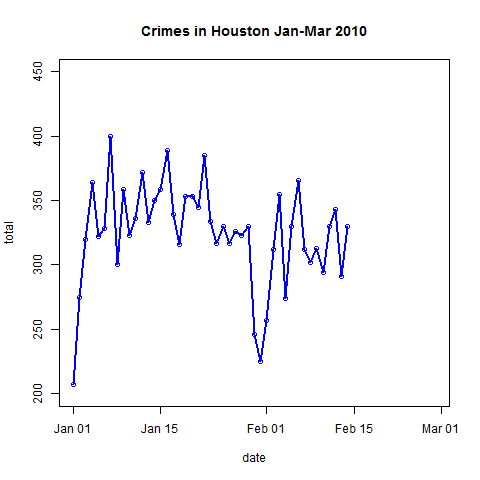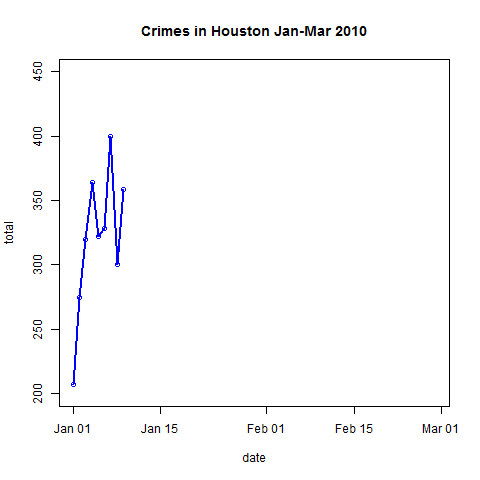
- Plot the data and explore trends
- Test hypothesizes with the data
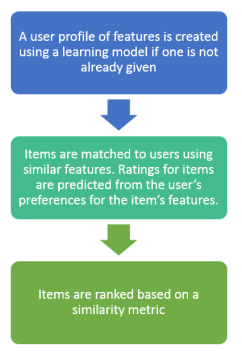 The user profile is typically based on historic user data to build a functional relationship representing how a user would rate a certain item.
The user profile is typically based on historic user data to build a functional relationship representing how a user would rate a certain item.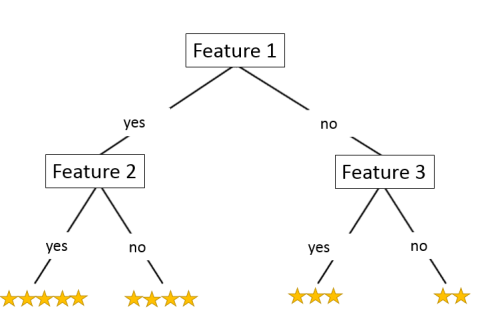
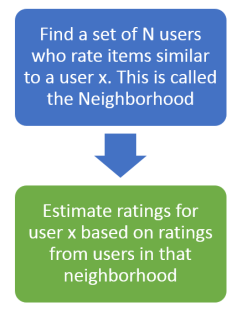 Cosine similarity or Pearson correlation are used to measure similarity between two users, and create the neighborhood of N users.
Cosine similarity or Pearson correlation are used to measure similarity between two users, and create the neighborhood of N users.